Home >> Products >> PrimerPlex >> xTAG
xTAG Technology
The xTAG technology developed by Luminex provides a method for the simultaneous detection of 150 different nucleotide sequences (or other analytes) in a single reaction. The technology makes use of two different types of microspheres - xTAG and MagPlex TAG.
An xTAG microsphere is a labeled 5.6 micron diameter polystyrene sphere. The labeling is done using two spectrally distinct fluorochromes. A MagPlex TAG microsphere is a superparamagnetic Carboxylated xMAP® Microsphere internally labeled with fluorescent dyes with magnetite encapsulated in a functional polymer outer coat containing surface carboxyl groups for covalent coupling of ligands.
The beads are pre-coupled with TAG oligonucleotide sequences. These TAG oligonucleotide sequences, also known as anti-tag sequences are complementary to the tags attached to the ASPE primers. Since each fluorochrome has a different intensity, 150 microsphere sets with specific spectral addresses are created for multiplex detection.
xTAG Challenges
Despite the target specificity of the xTAG and MagPlex TAG oligonucleotide sequences, chances are, that some of them may cross-react with custom designed oligos. Also, it is critical that no matter which beads are selected, if they are being multiplexed in one assay, they need to be from the same MFI group. Software that assists researchers in choosing the best TAG for each designed ASPE oligo can automate this process.
xTAG Mechanism
xTAG microspheres are interrogated individually in a fluid stream as they pass by two separate lasers in the MagPix, Luminex 200 or FlexMAP 3D analyzer. A 635-nm 10-mW red diode laser excites the two fluorochromes contained within the microspheres and a 532-nm, 13-mW yttrium aluminum garnet (YAG) laser excites the reporter fluorochrome (R-phycoerythrin, Alexa 532, or Cy3) bound to the microsphere surface. High-speed digital signal processing classifies the microspheres based on its spectral address and quantifies the reaction on the surface. Thousands of microspheres are interrogated per second by the signal detection platform, resulting in an analysis system capable of analyzing and reporting up to 100 different reactions in a single reaction vessel in just a few seconds per sample.
xTAG Chemistry |
|
1. Multiplex PCR: The region of interest is amplified using Multiplex PCR assays. A user can design multiplex PCR primers using PrimerPlex. |
 |
2. Exo/SAP Treatment of PCR Product: To remove the excess primers and oligonucleotide, the PCR reaction is treated. |
|
3. Multiplex ASPE: In this step, the PCR reaction is now subjected to a primer extension assay. It should be specific to the allele that is being analyzed (Allele Specific Primer Extension). The 5’ end of the ASPE primer is attached to a xTAG or MagPlex TAG universal tag sequence. ASPE primer design using PrimerPlex. |
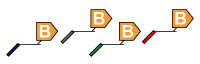 |
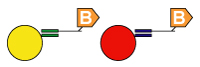
4. Universal Array Sorting: The hybridization of the 5' universal tag to the complementary anti-tag sequence takes place. The anti-tag sequence is coupled to a particular TAG bead set. |
 |
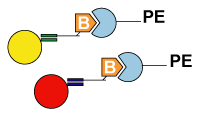
5. Detection: After hybridization, the xTAG or MagPlex TAG beads are read by the analyzer. Then the data analysis software analyzes the results. Applications1. High Throughput SNP genotyping |
 |


