Products >> SimMet® >> Features
Robust Relational Metabolite Database
The SimMet database includes 68,459 metabolite species from various biological sources such as Humans, E.coli. and supports 2,34,284 MS/MS spectra with 354 adducts for 9,390 metabolites from NIST MS/MS databases. Additional biological information such as metabolite common name, systematic name, mass, composition and links to the other databases are also made available for easy reference.
SimMet® offers accurate identification of metabolites by matching observed MS/MS spectra against standard MS/MS spectra of metabolites.
Support for Multiple File Formats
SimMet® accepts experimental mass spectrometry data in standard file formats such as .txt, .xls, .mzData and .mzXML. It can read Thermo Scientific (*.raw) and SCIEX (*.wiff) and Bruker Corporation's (.baf, .yep and .fid) native data files. The program is capable of importing data from complete chromatographic runs i.e up to 200,000 scans.
LC-MS and MS/MS Data Processing
SimMet® supports robust LC-MS data processing techniques such as peak detection, picking and identification. The data is automatically processed in batch mode to generate tables of detected metabolites for the retention time range specified. The tool automatically tabulates all the m/z values, abundance, isotope level, charge state and MS/MS data. SimMet® automatically aligns detected compounds with the corresponding MS/MS spectra for accurate metabolite identification based on exact mass search as well as precursor, product ions and spectral pattern matching.
SimMet® offers intuitive graphical interpretation of metabolite profiles via Pie Charts, Bar Charts and comparative analysis of metabolite profiles across samples using frequency tables.
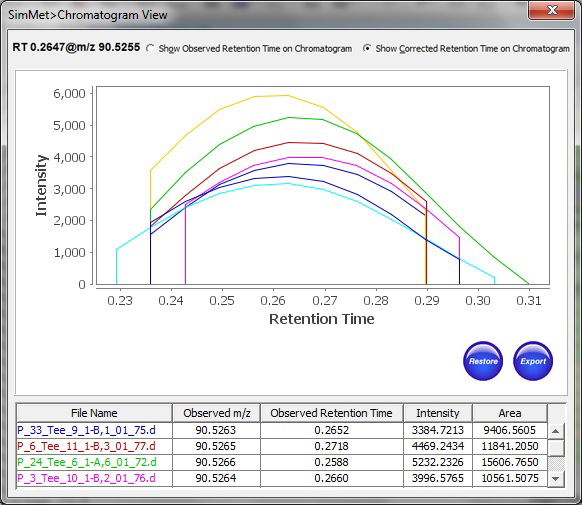
In addition, SimMet® aligns metabolites across biological samples based on Retention Time, Charge State, Probability, Score and Pattern Score to facilitates convenient investigation of data scattered across different scans or profiles. The program displays TIC (Total Ion Chromatogram), XIC (Extracted Ion Chromatogram) and BPC (Base Peak Chromatogram) for both MS and MS/MS levels. The chromatogram plots can be zoomed into, so as to investigate and highlight important peaks. The plots can be exported for sharing results in the lab.
High Throughput MS and MS/MS Data Analysis
SimMet® enables metabolite identification by matching observed metabolite precursor ions and product ions against known metabolite spectra/structures/fragments available in the SimMet® database. SimMet® offers an intuitive user interface to perform such analysis with ease besides displaying the identified 2D structure, fragment ions, annotated MS or MS/MS spectra and other information of identified metabolite in the same workspace.
SimMet® analyzes MS/MS data of 10,000 scans in a single search run. High resolution accurate mass data with error tolerance between 1-50 parts per million (ppm) and 0.1 to 2000 millidaltons (mDa) can be analyzed. The results are sorted using an innovative compound identification and spectral pattern matching algorithm that maps the degree of proximity of theoretical metabolites with experimental data.
Differential Metabolomics for Biological Samples
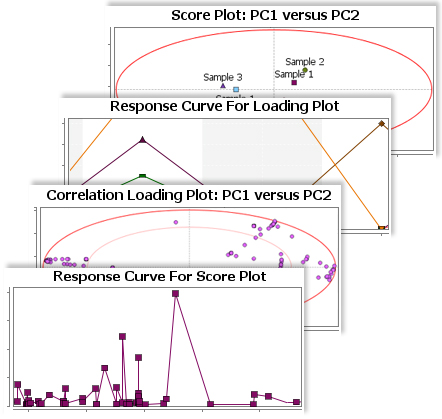
The Principal Component Analysis (PCA) for metabolites in SimMet® provides a visual representation of the relationship between biological samples and metabolites. Researchers can easily analyze similarities and differences amongst various biological samples. Principal Component Analysis (PCA) generates two 2-D plots, Scores plot and Loadings plot. In order to pinpoint probable biomarkers, the 2-D plots are supported with Confidence Ellipses such as Correlation Loadings and Hotelling's T2 ellipses.
Quantitative Metabolomics
SimMet® performs quantitative data analysis of metabolites present in samples using calibration curves constructed from dilution series and internal standards. SimMet® predicts the amount of analyte in samples using the fitted line plot methodology for regression.
Mass Spectra Annotation with Identified Metabolite
SimMet® can annotate mass spectra with the metabolites identified from MS and MS/MS data. Mirror plot is also available for showing observed vs standard spectra of the identified metabolites.This helps in interpreting mass spectra by highlighting the experimental m/z values that match those of theoretical metabolite structures from the product database. Each matched peak can be annotated with the metabolite fragment. The annotated spectra can be exported as an image (.JPEG or .PNG) to MS PowerPoint to facilitate information sharing amongst research groups. The annotated mass spectra can be re-sized to fit to a page. Alternatively, users can zoom in to or out of the plot by specifying the m/z range or by selecting the range on the plot and exporting the results.
*NIST spectral library is accessible upon product purchase.


