Home >> Products >> SimLipid >> Waters® TQ-S MPIS/NLS-based Shotgun Lipidomics Methods
Waters® TQ-S MPIS/NLS-based Shotgun Lipidomics Methods
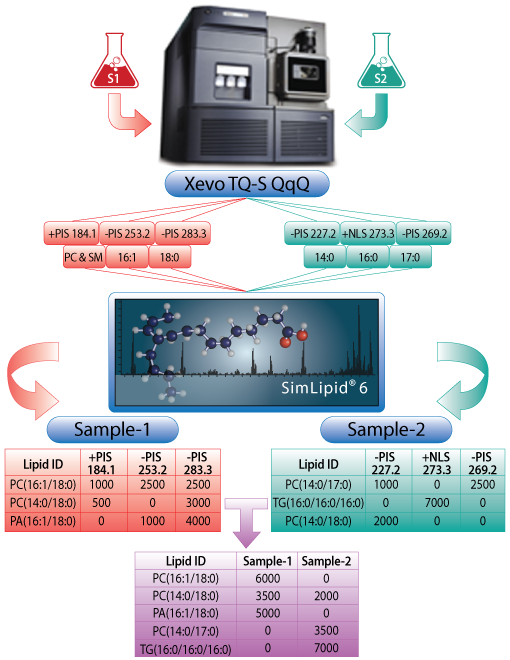
Automating lipid identification and quantitation using data from Waters® Xevo® TQ-S MPIS/NLS-based shotgun lipidomics methods
- Database containing 40,312 lipid structures.
- 5211 set masses (transitions) of PIS/NLS experiments for fatty acid chains, and head groups of lipid species from different classes and sub-classes.
- Identify target lipids based on exact mass database search, and the target headgroup, or fatty acid chain corresponding to the set masses of the PIS/NLS experiments.
- Isotope peak correction to facilitate accurate quantitation of lipids.
- Align lipids across biological samples based on short name e.g., PC(28:2); name without double bond positions e.g., PC(18:1/18:1) instead of PC(18:1 (9Z)/18:1(9Z)).
- Calculate normalized response of analytes w.r.t. the response of the internal standards that belong to the same class/subclass.
- Heatmap, Concentration X Composition reports of lipid classes, and fatty acid chains.
- Additional lipid ontology based filters, e.g., only even/odd numbers of double bonds, and carbon (C) atoms in the fatty acid chains of target lipids.