Fragmentation Techniques for Mass Spectrometry based Glycopeptide Profiling
June 07, 2022
Mass spectrometry has emerged as a powerful tool for glycoprotein analysis. With the advent of tandem mass spectrometry, which provides highly informative glycan and peptide fragments, the structural elucidation of glycoproteins has become a lot easier. The present generation mass spectrometry instruments are equipped with various fragmentation techniques for the comprehensive characterization of glycoproteins. These techniques have a vital role in identifying the modified glycan residue and characterizing the N-glycan on a peptide by generating glycan-specific fragment ions. The nature of glycan moieties is versatile, and with the removal of glycans from the peptides, it is easy to match the resultant fragments to a theoretical database using a standard data analysis workflow. Therefore, the fragmentation strategies have significant importance in the mass spectrometry-based glycoproteomic analysis. This blog discusses four different fragmentation strategies, namely CID, HCD, ETD, and UVPD, in context with efficient glycopeptide profiling.
Collision-induced dissociation (CID):
Collision-induced dissociation (CID) is also known as collisionally activated dissociation (CAD). CID is the most commonly used fragmentation method for glycoproteomic studies. In this method, the molecules (peptides/protein) get accelerated by colliding with a neutral gas (e.g., nitrogen), which breaks the molecular bond and generates tandem mass spectra. The resulting fragments are called B- and Y-type ions from the glycan portion of a glycopeptide conjugate. The CID fragmentation is of two types- (a) trap-type (derived from an ion trap instrument) and (b) beam-type (derived from a quadrupole-time-of-flight instrument). For small and low-charged peptides, the CID fragmentation is more effective. However, CID is not a suitable methodology for fragmenting intact proteins and peptides with labile glycosylation patterns.
Electron-transfer dissociation (ETD):
Electron-transfer dissociation (ETD) method is advantageous over the CID technique, as it caters to the above-mentioned limitations by fragmenting longer peptides or even the complete proteins. In this method, the multiply-charged gaseous macromolecules are subjected to fragmentation in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). ETD spectrum validates the peptide sequence as the technique is crucial for fragmenting the peptide backbone to yield the z- and a majority of c-ions. The ETD spectra clearly distinguish the residue (Asn, N) where the glycan structure is attached. The application of the ETD fragmentation method in proteomics is mainly associated with the analysis of PTMs since fragment ions tend to retain the modifications that are often lost with CID approaches.
High-energy collision dissociation (HCD):
A CID technique specific to the orbitrap mass spectrometer is known as the beam-type CID, higher-energy C-trap dissociation, or high-energy collision dissociation (HCD). Here the fragmentation occurs external to the trap. While comparing it with traditional ion trap CID, the HCD fragmentation method offers higher activation energy and shorter activation time. The HCD fragmentation method generates B- and Y-type fragment ions similar to CID. The y-ions tend to show dominance, whereas b-ions are further fragmented into a-ions or smaller species due to higher energy.
Ultraviolet Photodissociation (UVPD):
UVPD is an innovative fragmentation technique where high-energy photons are used to energize the ions, cleave the bonds, and produce fragment ions. UVPD generates glycosidic cross-ring fragments of glycans resulting in A/X fragments that are important for the characterization of intersaccharide linkages in complex glycosylated molecules. An HCD-triggered UVPD MS approach can be ideal for comprehensive glycopeptide profiling.
The studies suggest that CID and HCD fragmentation methods are suitable for glycopeptide identification, whereas the ETD method interprets the glycosylation sites by maintaining the glycan-peptide linkage. For the compounds with the higher charge states, the ETD method provides better-fragmented spectra than CID or HCD. However, the major disadvantage of ETD is the longer acquisition time. A hybrid MS/MS method, Electron-transfer higher-energy collisional dissociation (EthcD), is also found effective for glycoproteomics analysis. This technique combines CAD and ETD to generate meaningful glycan and peptide fragments with considerable abundance.
These fragmentation techniques generate an enormous amount of glycan (glycosidic and cross ring) and peptide (N-terminal and C-terminal) fragments. Interpreting these fragments manually to construct an accurate glycopeptide structure is challenging. SimGlycan® software accommodates a robust in-silico fragment database and the capability of generating structure-specific theoretical ions using a proprietary algorithm. For analyzing the experimental released glycan and glycopeptide spectra, the software allows you to specify the type of fragments (glycan and peptides) based on the fragmentation technique employed. To know more about SimGlycan software, book a meeting with us.
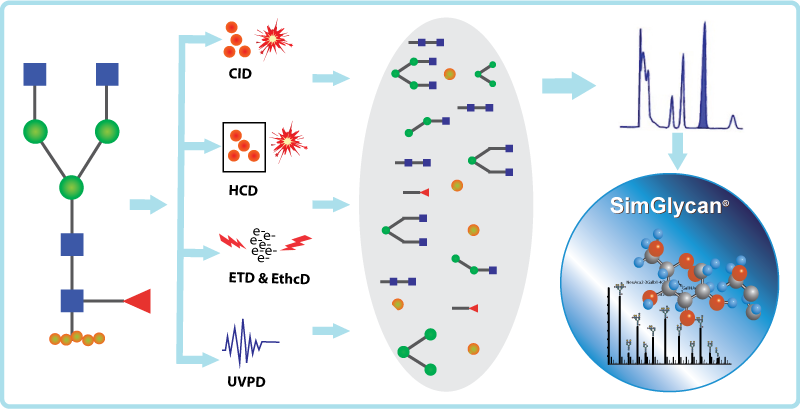
Fig: Various types of fragmentation techniques supported in SimGlycan software.
| Comment | Share |
|


