Fast and Accurate Lipid Profiling of Serum Samples by Probe-Electrospray Q-TOF MS
October 20, 2022
The utilization of liquid chromatography-based mass spectrometry (LC-MS) has been the gold standard method for high-throughput lipid profiling. The limitation of this approach is extensive sample preparation, long analysis time, and system contamination. With the advent of probe-electrospray ionization (PESI), a novel ambient ionization technique, these limitations were overcome, and lipid analysis can be performed directly and rapidly using biological samples. This direct injection method reduces the total experiment time significantly, as near-crude samples can be subjected directly to MS analysis that finishes within seconds. Here we present a proof-of-principle experiment to demonstrate the significance of this novel workflow in ultrafast and direct lipid profiling.
A serum sample with no clinical implications was purchased from Kohjin Bio (Japan). A 50 μL aliquot of serum was diluted with 450 μL water and subsequently mixed with 500 μL ethanol. The mixture was thoroughly vortexed for 30 sec. A portion of this sample was transferred to a sample loading cartridge and measured directly with the DPiMS-QT; a Q-TOF MS with probe-ESI interface (Shimadzu Corporation) in MS and MS/MS modes. The ionization outage time, sample outage time, and sample position were set at 180 milliseconds, 30 milliseconds, and -46.00 mm, respectively. For MS/MS scans, the collision energy was ramped from 30-50 V across the 100-850 m/z range. SimLipid software (PREMIER Biosoft, USA) was used for performing data analysis.
We performed the MS analysis on the serum sample and the MS data files were directly loaded into SimLipid software for lipid identification. A high-throughput lipid search using 5 ppm precursor ion error tolerance was performed across the SimLipid database comprising over 40,000 lipids. The characteristic five PC lipids i.e., PC (18:2) observed at 518.3252 m/z, PC (16:0/20:4) observed at 780.5549 m/z, PC (16:0/20:3) observed at 782.5705 m/z, PC (16:0/22:4) observed at 808.5862 m/z, PC (18:0/22:6) observed at 832.5862 m/z were identified in the serum sample based on the exact mass search approach for MS analysis. For accurate identification, the MS/MS data were further subjected to high-throughput lipid MS/MS search using precursor and product ion error tolerance of 5 ppm. The accuracy of the identified lipids was ensured using an innovative ranking algorithm that relies on the percentage match between the experimental lipid fragments and the theoretical fragments. Results of both MS and MS/MS analysis are exported into Microsoft excel files along with detailed information such as lipid class, category, lipid IDs, and matched fragment ions (for MS/MS data). This proof of concept novel method for lipid identification using the PESI-QTOF-MS was conducted.Further details are available in the poster.
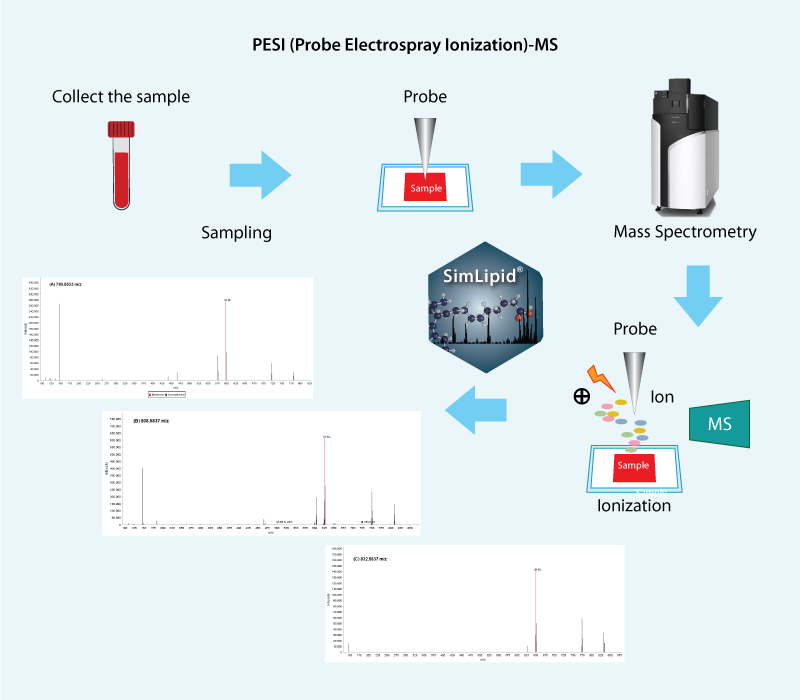
Fig: Lipid identification workflow for PESI-QTOF-MS using SimLipid software
| Comment | Share |
|


