Home >> Blog >> Identification of N-Glycans from Cetuximab (mAb) using LC/MS analysis with SimGlycan Software
Rapid and Accurate Identification of Cetuximab N-Glycans using UHPLC Coupled with Orbitrap-Based MS with SimGlycan Software
May 06, 2020
Cetuximab, a chimeric IgG1 monoclonal antibody (mAb), is used in the treatment of head and neck cancer, and colorectal cancer. Structural characteristics of cetuximab play a key role in its therapeutic application. Currently, liquid chromatography (LC) coupled with tandem mass spectrometry (MS/MS) technique is the gold standard method for glycan characterization of mAbs. However, it needs a robust bioinformatics solution that can process large sets of data generated using this method and annotate the glycan structures accurately. SimGlycan software v 5.93 facilitates LC-MS data processing (which includes peak detection and picking, molecular feature finding, accurate clustering of MS/MS scans to compounds), and automated identification of glycan compositional structures by annotating in-silico fragment ions on peaks of the MS2 spectra.
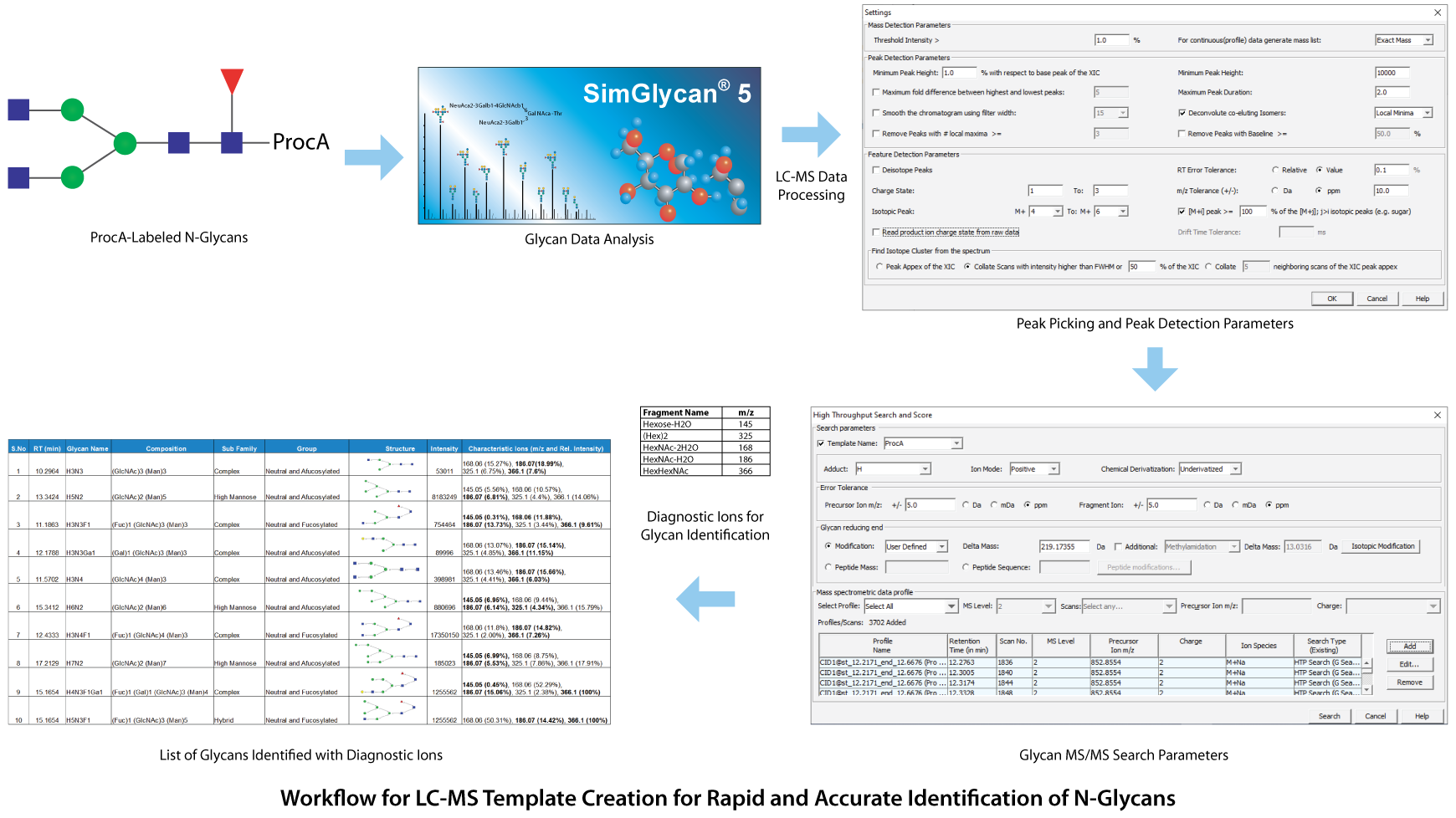
For performing rapid and accurate identification of N-glycans, a template-driven method was created. A glycan template storing curated glycan structures along with corresponding retention times, and other chromatographic details is created using the SimGlycan software. This facilitates rapid and accurate identification of glycans and their isomers by performing MS/MS database search using precursor m/z values, corresponding ion species, and retention times as initial search predicates. As a proof of concept, we created an LC-MS template for the identification of procainamide (procA) labeled N-glycans from cetuximab using UHPLC with a Thermo Scientific™ Accucore™ 150 Amide HILIC column (2.6 µm, 2.1 x 150 mm) coupled with Thermo Scientific™ Q Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer controlled by Thermo Scientific™ Chromeleon™ Chromatography Data System (CDS) Software.
The raw data was subjected to LC-MS peak picking and peak detection using an m/z tolerance of 0.011 Da and 0.1 mins for the minimum time span for the LC-peak. Then the LC compounds were analyzed using the MS/MS database using 5 ppm error tolerance for precursor and product ions. For each identified MS/MS spectrum, the possible glycan structures were obtained and these structures were validated based on their characteristic ions corresponding to oxonium and fragment ions. Similarly, the other glycan structures were assigned to different MS/MS spectra (see below image). This template-driven approach has proved to be effective in rapid and accurate identification of N-glycans in Cetuximab. For more details, please visit: http://premierbiosoft.com/citations/glycomics-posters.html
| Comment | Share |
|


