Data Independent Acquisition Methods in Lipidomics

August 31, 2021
Lipids, one of the key components of the biological processes, play a critical role in various biochemical and signalling pathways. Structural characterization of these lipids can provide insights about their importance in various bioprocesses. With recent advancements in hardware and software technologies, mass spectrometry (MS) has emerged as the frontline approach for both untargeted and targeted lipidomics studies. Data dependent acquisition (DDA) mass spectrometry has been the go to approach for lipidomics wherein precursor ions that produced the most abundant MS are selected and only these precursors are subjected to MS/MS. However, the major setback for this acquisition mode has been that the detection of low abundant lipids is relied on a hit-and-miss basis.
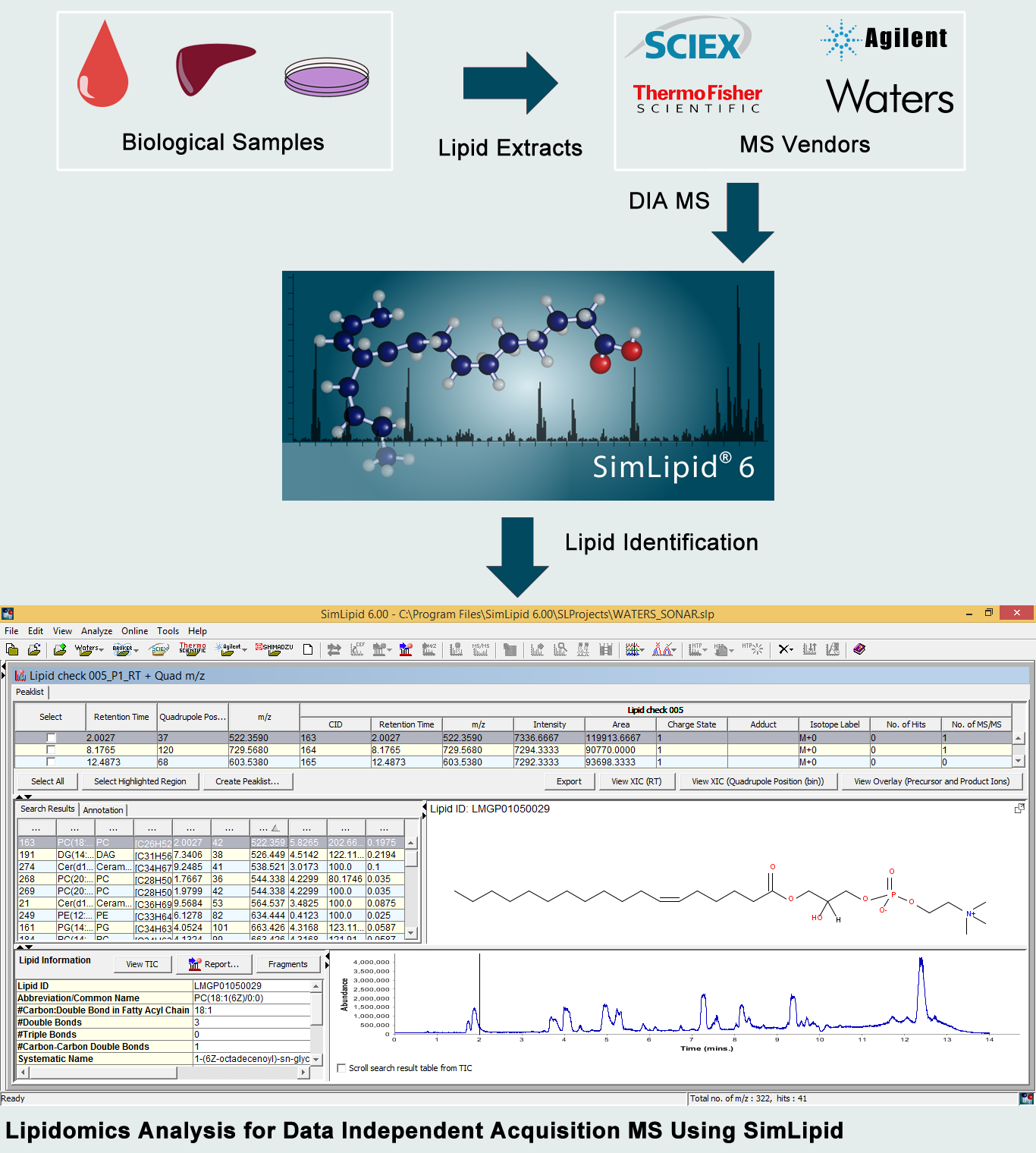
To overcome this challenge, data independent acquisition (DIA) has been developed wherein all the precursor ions within a time cycle were subjected to fragmentation in the mass spectrometer. This detection provides an unambiguous identification and quantification of all the lipids (both low and high abundant peaks). DIA MS has been supported by most MS vendors-Water-SONAR/HDMSE, Bruker (timsTOF), Agilent, Thermo (All ion fragmentation-AIF), SCIEX (MS/MSALL). Depending on the type of MS instrument vendors, the DIA strategies are divided into two: (i) Waters MSE, Thermo-AIF, SCIEX-MS/MSALL (ii)Waters SONAR, SCIEX-SWATH MS.
Waters DIA Method
The ion mobility (IM) separation feature in Waters instruments helps to generate the DIA based data. For SONAR/HDMSE workflow, two parallel alternating scans at two collision energies (low and high) are applied for MS/MS. For SONAR, a scanning quadrupole isolation window is applied and all the precursor ions in this window will be subjected to MS/MS, however for MSE workflow, the quadrupole window is not applicable. Both approaches will enhance the sensitivity of the fragment ions and also maximize the instrument duty cycle. In comparison to the DDA approach, there is no necessity of switching between the MS and MS/MS mode.
Bruker DIA Method
The use of Bruker’s trapped ion mobility spectrometry (TIMS) and parallel accumulation-serial fragmentation (PASEF) can assist with the in-depth lipidomics analysis.
Agilent DIA Method
To minimize the use of two or more chromatographic separations for obtaining the complete lipid profiling, Agilent has coupled the supercritical fluid chromatography (SFC) with a high resolution MS instrument. This comprehensive approach can provide the information corresponding to both polarities and help in generating a near-complete lipid profile.
Thermo DIA Method
The application of flow injection analysis (FIA) coupled with Thermo Orbitrap instruments helps to perform lipidomics studies by subjecting all the ions to MS/MS. This provides a high throughput lipid identification by utilizing the DIA based MS/MS acquisitions.
SCIEX DIA Method
The SCIEX's SWATH/MS/MSALL approaches are the two DIA strategies that are commonly used for lipidomics. In comparison of both approaches, for SWATH acquisition, an additional isolation window of 20-50 Da is applied across the m/z range prior to fragmentation.
Here are some of the challenges that DIA-based data analysis possesses:
- Size of the datafiles acquired using DIA MS are large and require large processing times.
- Data complexity increases when LC is involved
- Ion mobility feature (Waters MS instruments) will present an additional level of data complexity
- Lipid identification
Large Sizes of Datafiles
The DIA-based MS approach in comparison to the DDA method comprises larger data files due to the inclusion of all precursor ions and also due to time-cycle based acquisition. Data loading of these DIA files takes longer times.
LC-MS Data Processing
The addition of LC-MS will increase the complexity of the data for data analysis. Also, to generate the LC peaks, the time-cycle based acquisition parameters has to be taken into account.
Ion Mobility
The ion mobility dimension will further increase the complexity of the data analysis. The parameters from ion mobility like quadrupole bins/drift time are to be included in the peaklist generation.
Lipid Identification
The peaklist with additional information (LC-MS, ion mobility) is subjected to a lipid search and this will have a significant impact on the identification and also will require longer analysis times for lipid identification.
SimLipid with its built-in automated data loading ability for multiple MS vendor-specific files can dramatically decrease the loading times for DIA data. With robust peak picking and peak detection algorithms for LC MS data, the data preprocessing time can be significantly reduced using SimLipid. A high throughput lipid search can be performed across a robust lipid database comprising over 40,000 lipid profiles. Also, the MS/MS spectra can be annotated with lipid IDes based on the lipid fragment matches.
| Comment | Share |
|


