Home >> Blog >> A High Throughput Qualitative and Quantitative Lipidomics Workflow using FIA-HRMS with Tandem MS (DIA) Based Method with SimLipid Software
A High Throughput Qualitative and Quantitative Lipidomics Workflow using FIA-HRMS with Tandem MS (DIA) Based Method with SimLipid Software
September 09, 2019
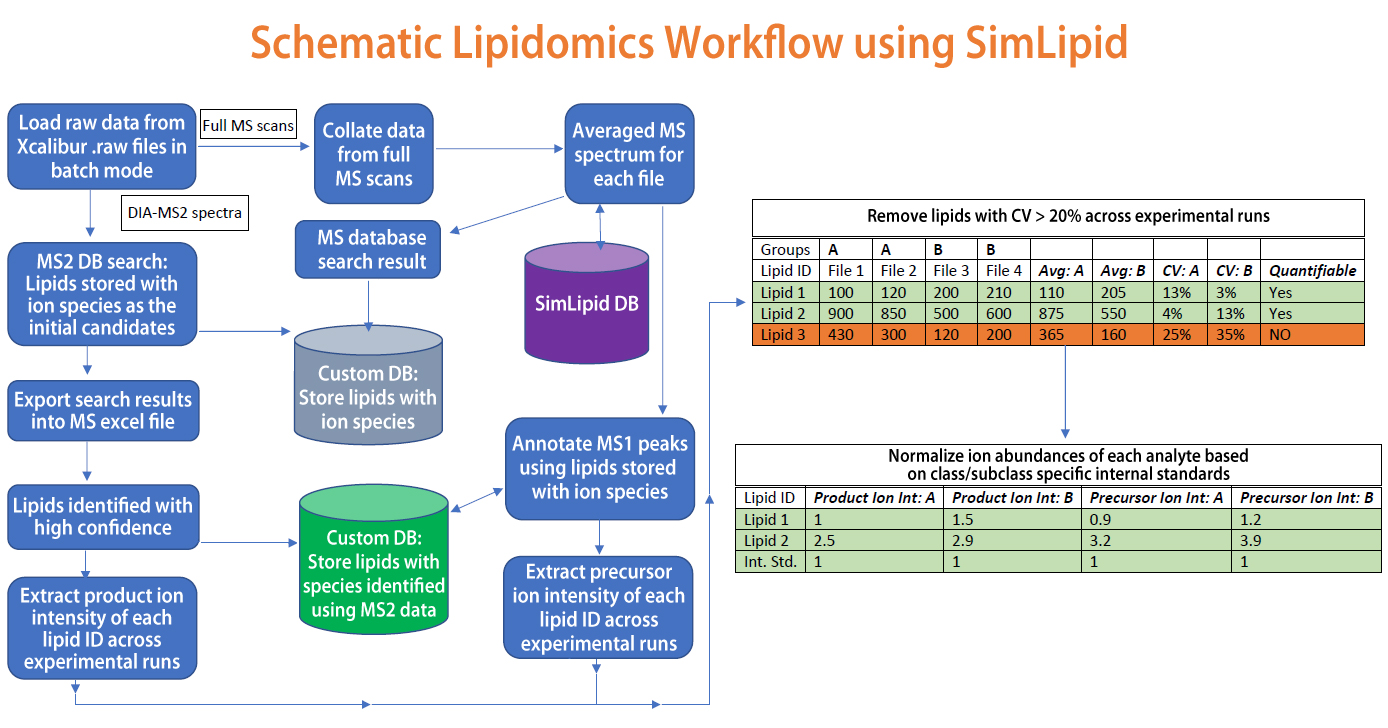
Application of flow injection analysis (FIA) coupled with high-resolution mass spectrometry (HRMS) is currently trending as a suitable method for lipidomics study. In this method, FIA coupled with Thermo Orbitrap MS instrument assists in measuring data-independent acquisition (DIA) based MS/MS data for lipid species. MS and MS/MS data acquired were subjected to data processing, lipid identification, and quantitation using SimLipid software. Averaged MS data were extracted from the raw MS data during loading with a peak m/z tolerance of 5ppm. A selective lipid database search with 5ppm mass tolerance was performed. The results obtained were converted into a custom database (DB MS1) and imported to the SimLipid server database program. The MS/MS data were subjected to a search on the custom database (DB MS1). From these results, the unlikely lipid ion species were removed manually and a total of 592 unique lipids were identified with high confidence. These unique lipids were exported to Microsoft Excel and total ion abundances for the product ions were extracted across the experimental runs. The coefficient of variation (CV) was calculated for each lipid species using in-built Excel formulas and only those lipid ions (330 unique) with CV<20% were selected. The product ion abundances along with their respective parent lipid ion species were stored as another custom database (DB MS2). The averaged MS spectra were re-annotated using the DB MS2 custom database and 206 unique lipids with a CV < 20% of precursor ion abundances across replicates were obtained. The ion abundances of the class-specific internal standard were used for quantitation. Also, a comparative analysis was performed across study groups using normalized precursor versus product ion abundances. Both cases, wherein lipid species that showed similar and different patterns in terms of increase/decrease in normalized precursor ion abundance versus the normalized product ion abundances were compared. For more details, please visit http://premierbiosoft.com/citations/lipidomics-posters-technical-application-notes.html.
| Comment | Share |
|


